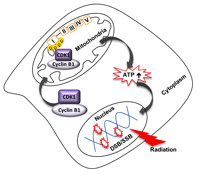
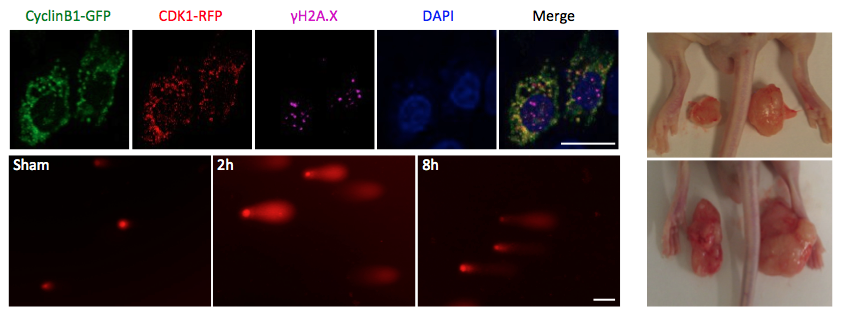
Further, mitochondria targeting expression of kinase-deficient mutant CDK1 significantly reverses radiation-enhanced mitochondrial ATP generation and inhibits DNA repair; overexpression of CDK1 phosphorylation-deficient mitochondrial respiratory chain complex I (CI) subunits compromises radiation-enhanced mitochondria ATP generation. These results indicate a novel mechanism regulating communication between mitochondrial bioenergetics and nuclear DNA damage repair under genotoxic stress conditions, by which mitochondria-relocated CDK1 boosts ATP generation to meet the increased fuel demand for DNA repair.
Novel anti-cancer strategies targeting abnormal cellular energy metabolism in cancer cells
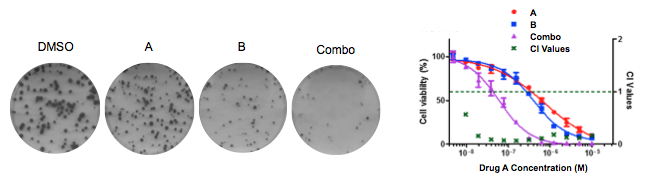
The bromodomain and extra-terminal domain (BET) family proteins are epigenetic readers for acetylated histone marks. Emerging BET bromodomain inhibitors have exhibited antineoplastic activities in a wide range of human cancers through suppression of oncogenic transcription factors, making it a promising drug candidate, particularly for cancers driven by aberrantly activated oncogenic transcription programs. However, the preclinical activities of BET inhibitors in advanced solid cancers are moderate at best, raising concerns over their clinical potential as monotherapy. To improve BET-targeted therapy, we interrogated mechanisms mediating antineoplastic activity and resistance to BET inhibitors in cancer. We found that BET inhibition suppresses glycolysis through inhibiting the glucose uptake and consumption, and combination of BET inhibition and glutaminase inhibition has significant synergistic effect in KRAS-mut lung cancer and colorectal cancer cells. Also we found that the MAPK pathway contributes to the intrinsic resistance to BET inhibitors in colorectal cancer cells. These findings propose potential combination strategy of BET inhibition with inhibition of Glutaminolysis or MAPK pathway to optimize BET bromodomain-targeted therapy of cancer.

Cellular radiation responses and radiation resistance in cancer therapy
Mammalian cells are able to induce an adaptive response under certain genotoxic stress conditions including ionizing radiation to enhance their survival. DNA damage is one of the major radiation-induced cell events, while DNA repair is necessary for cell's survival. It is well known that DNA repair is an energy-consuming process, which indicates that mitochondria is possibly involved in radiation-induced DNA damage repair.
My recent study found that sublethal dose (5 Gy) of radiation enhances mitochondrial ATP generation in normal cells, which is paralleled with enhanced mitochondrial relocation of cell cycle kinase CDK1 and nuclear DNA repair capacity.
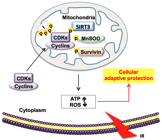
Low dose radiation Hyperradiosensitity (HRS) and Increased Radioresistance (IRR) are typical cellular adaptive radiation responses observed in most cancer cells, which caused a higher radioresisance in cancer radiotherapy following guidance of CT scan. In my recent study, Cyclin D1/CDK4 is found relocated in mitochondria under low dose ionizing radiation, where it phosphorylates MnSOD, causing enhanced MnSOD activity and mitochondrial respiration, allowing cells to sense environmental genotoxic stress to improve cell survival. This finding shed light on the mechanisms underlying cellular adaptive protection under oxidative stress conditions and will help to design the novel approaches to target recurrence of radioresistant tumor and protect normal cells from radiation-associated injury.
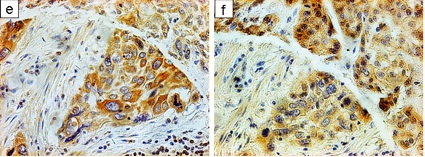
Accumulating evidence indicated that cancer stem cells (CSCs) contribute to tumor radioresistance. Our recent work established fractionated ionizing radiation (FIR)-surviving cells from multiple types of tumor cells. This FIR-surviving fraction represents stem cell and aggressiveness features, suggesting its role in cancer radioresistance and relapse. RNA-seq data showed that a group of genes encoding mitochondrial proteins implicated in mitochondrial energy metabolism is upregulated in FIR-surviving breast cancer stem cells (BCSCs), suggesting the possibility of the enhanced mitochondrial energy production in these cells.
Actually, We found that mitochondrial ATP generation is altered in the FIR-surviving cells compared to their parental cells, and that another single dose of sublethal dose of radiation increased the mitochondrial ATP level in FIR-surviving colon cancer cells, providing evidence that the FIR-surviving stem-like cancer cells may also shift their energy production from glycolysis to mitochondrial oxidative phosphorylation. The elucidation of the mechanisms underlying the regulation of mitochondrial energy metabolism of these radioresistant cells will help to develop the new strategies and targets to improve the outcome of cancer radiation therapy.
Contributions:
- Measurement of mitochondrial function in in vitro cells and in vivo fresh tissues including oxygen consumption, mitochondrial ATP generation, mitochondrial superoxide level and MnSOD activity to assess the mitochondrial homeostasis.
- Foci formation assay, comet assay and immunoblot to test DNA damage and repair.
- Preparation of subcellular fractions including mitochondria, cytosol and nuclear from cultured cells and fresh animal tissues.
- Immunofluorescence and immunohistonchemistry assay.
- Tumor sphere formation, clonogenic survival, cell invasion and gap filling assay.
- Mouse xenograft tumor growth and treatment.
- ChIP-PCR
- Real-time PCR
- Cell culture, DNA cloning and cell transfection.

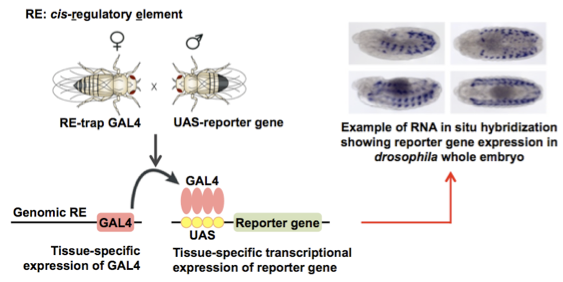
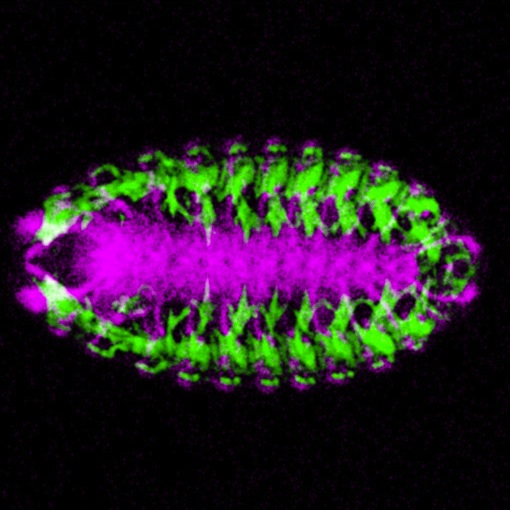
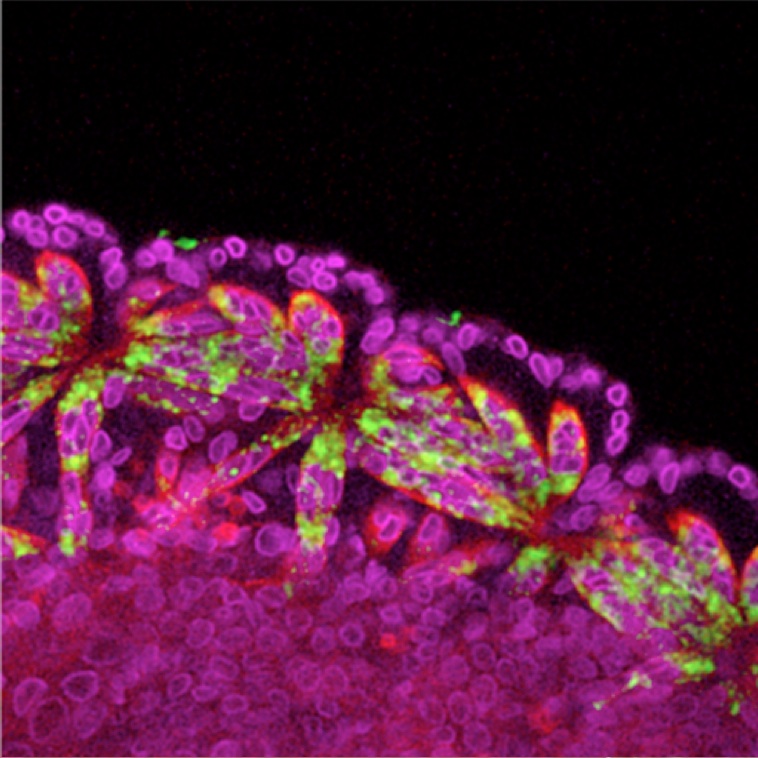
Identification of cis-regulatory elements of nautilus gene in Drosophila
A cis-regulatory element is a region of non-coding DNA sequence in or near a gene required for proper spatiotemporal expression of that gene, often containing binding sites for transcription factors. We identified cis-regulatory elements of nautilus gene in Drosophila as a start to study how this gene's expression is regulated. Using a comparative genomics tool, EvoPrinter, I identified 11 candidate cis-regulatory elements of nautilus gene, which are evolutionarily conserved in 12 Drosophila genomes.
Using Gal4-UAS reporter system and RNA in situ hybridization in Drosophila embryos, I verified the regulatory function of these candidates on nautilus gene expression and found that 6 of total 11 candidate cis-regulatory elements are involved in the regulation of nautilus gene expression and muscle development in different spacial and temporal manners.
Contributions:
- Establishment of transgenic fly lines
- Preparation of total RNA and genomic DNA from Drosophila adult, pupae and larvae
- Prediction of cis-regulatory elements using bioinformatics tools (BLAT, EvoPrinter)
- Gal4-UAS reporter gene system and RNA in situ hybridization in Drosophila embryos
- DNA cloning
- Southern blot
- ChIP-PCR
- Real-time PCR
- Luciferase reporter assay

Transgenic fly line for functional study of nautilus gene in Drosophila muscle development
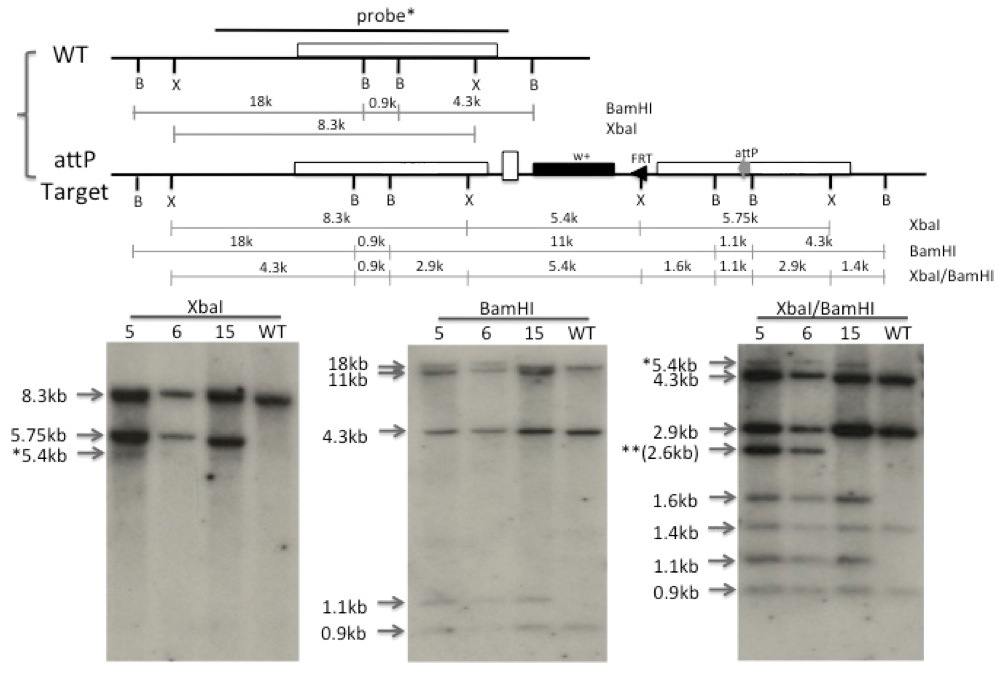
Gene targeting is a genetic technique that uses homologous recombination to change an endogenous gene.Ends-in makes use of p-element transposon insertion, FLP-FRT site-specific recombination system, I-SceI rare-cutting endonuclease mediated DSB and homologous recombination (HR) repair system to target desired mutation into the interest locus in the genome (Rong, Y. Science, 288, 2013-2018, 2000). PhiC31 integrase mediates sequence-directed, irreversible and highly efficient integration between a bacterial attachment site (attB) and a phage attachment site (attP). SIRT combines phiC31-mediated site-specific integration and ends-in targeting to generate mutations of a gene at its endogenous locus. An attP site is firstly targeted to the interest locus through ends-in targeting and subsequent reduction. Further modifications of the same locus are introduced by integration of an attB-containing plasmid with a modified target locus. Thereafter, a series of modification of the same target locus can be introduced repeatedly with once attP placement step. It is a laber- and time-saving tool that allows repeated modifications of a gene, including single amino acid replacements, small domain deletion and complete deletion (Gao, G. PNAS, 105(37), 13999 –14004, 2008).
Using SIRT, I established a transgenic fly line with an attP site inserted into nautilus gene in Drosophila genome, which makes it easier to study the functions of nautilus gene by repeatedly introducing a series of modifications in a time- and labor-saving way. It has been used in a study for deletion of entire 3’UTR region and mutations of some candidate miRNA binding sites to verify the targeting effect of series of miRNAs on nautilus gene.In addition, since no commercial ChIP grade antibody to nautilus available, introducing a tag to nautilus using this fly line will make it possible to screening nautilus target genes using anti-tag antibody

Aurora-A interacts with Cyclin B1 and play roles in tumorigenesis
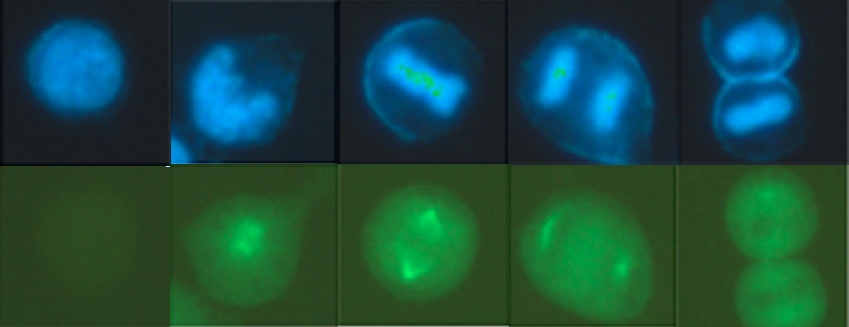
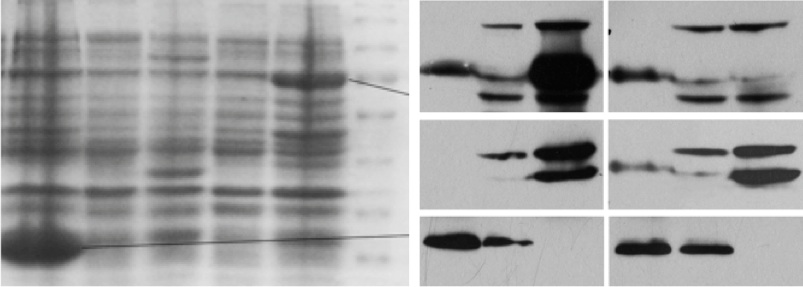
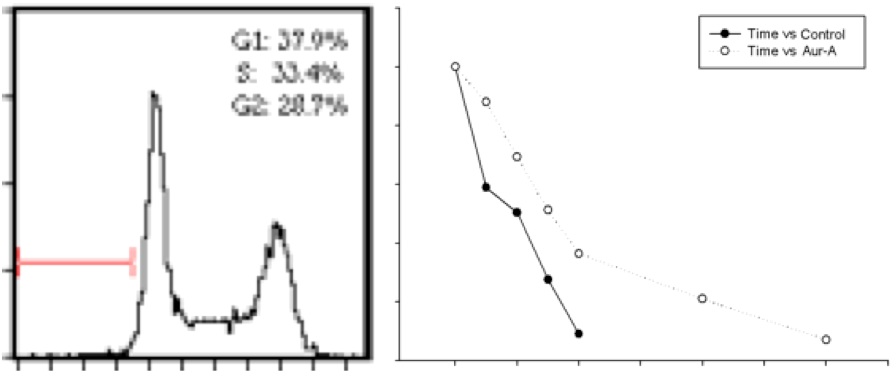
Aurora-A, as a cell cycle regulator, has been reported overexpressed in many types of cancers, leading to centrosome amplification and aneuploidy, which can be a driving cause of genomic instability and tumorigenesis. I found that overexpression of Aurora-A delays Cyclin B1 degradation, resulting in an increased Cyclin B1 protein level; while Aurora- A RNAi enhances Cyclin B1 degradation. Further study showed that Aurora-A interacts with Cyclin B1 and inhibits ubiquitin-mediated degradation of Cyclin B1, which depends on Aurora-A kinase activity. These results provide new insight into the mechanism of how deregulated Aurora-A contributes to genomic instability and carcinogenesis.
Contributions:
- Establishment of Aurora-A overexpression and knockdown ESCC cell model
- Immunohistonchemistry and immunofluroscence assay
- Preparation of total RNA from cells and performance of RT-PCR
- Preparation of protein sample and performance of immunoblotting
- Immunoprecipitation and pull-down assay to analyze protein interaction
- Cell culture, DNA cloning and cell transfection

Production of polyclonal and monoclonal antibody against cardiac troponin-I (cTn-I)
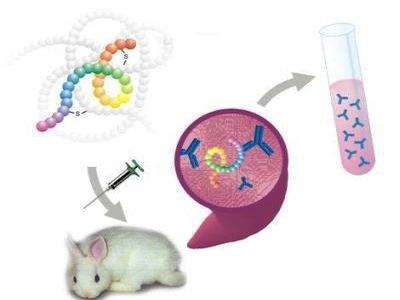
Antibodies are host proteins that are produced by the immune system in response to foreign molecules that enter the body. These foreign molecules are called antigens, and their molecular recognition by the immune system results in selective production of antibodies that are able to bind the specific antigen. Antibodies are made by B-lymphocytes and circulate throughout the blood and lymph where they bind to their specific antigen, enabling it to be cleared from circulation. This ability of animal immune systems to produce antibodies capable of binding specifically to antigens can be harnessed to manufacture probes for detection of molecules of interest in a variety of research and diagnostic applications.
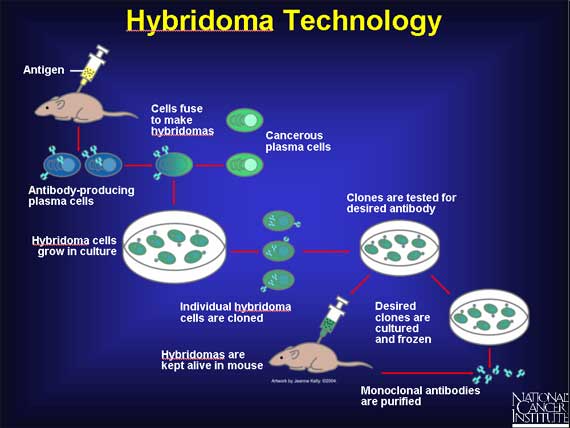
Antibody production involves preparation of antigen samples and their safe injection into laboratory or farm animals so as to evoke high expression levels of antigen-specific antibodies in the serum, which can then be recovered from the animal. Polyclonal antibodies are recovered directly from serum (bleeds). Monoclonal antibodies are produced by fusing antibody-secreting spleen cells from immunized mice with immortal myeloma cell to create monoclonal hybridoma cell lines that express the specific antibody and secrete into cell culture supernatant.
Using cTn-I protein isolated from patient heart or commercially synthesized cTn-I polypeptied, we produced high titer rabbit polyclonal antibodies and mouse monoclonal antibodies against human cTn-I, which can be used in lab research or clinical diagnosis.
Contributions:
- Animal immunolization though i.p. injection
- Collect polyclonal antibody (serum) from immunolized rabbits
- Isolate spleen cells from immunolized mice and culture monoclonal hybridoma cell lines
- Collect specific antibody from cell culture supernatant
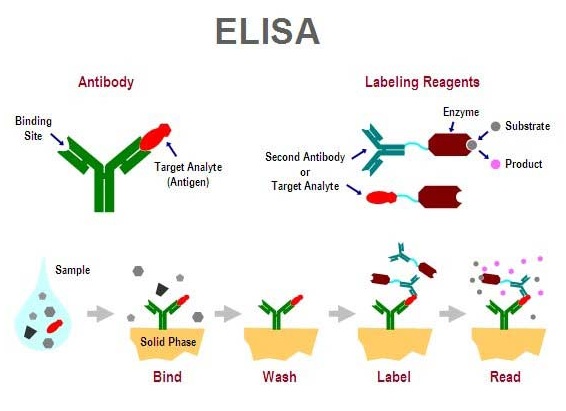
- Test antibody titer using enzyme-linked immunosorbent assay (ELISA)